Summary
Living cells have a fascinating ability to generate complex and dynamic internal structures. Nowhere is this property more evident than during mitosis and cytokinesis: in a very short time (often of the order of a few minutes) cells alter their shape, duplicate and partition their internal components, and divide into two apparently identical halves. These dramatic morphological changes need to be carefully coordinated with each other in space and time. To learn more about the principles underlying this coordination, we focus on the events at the end of the cell cycle: chromosome segregation and cytokinesis, in the yeast Saccharomyces cerevisiae. Regulatory systems identified in yeast are then validated in animal cells (such as Drosophila), to ensure that our key findings are relevant for the fidelity of mitosis and genetic stability in multicellular organisms.
Research projects
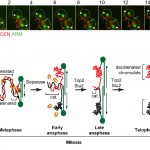
- We are interested in elucidating the fundamental principles of mitotic chromosome structure, and how this is regulated over the cell cycle. In other words, what gives mitotic chromosomes their shape, and how is their structure modified during their segregation in mitosis? We discovered that in yeast, replicated chromosomes arms are intertwined until the last step of mitosis, known as anaphase (Titos et al., “Chromosome length and perinuclear attachment constrain resolution of DNA intertwines”, J Cell Biol 2014, 206: 719–733). Removal of these DNA entanglements is accomplished by the anaphase activities of DNA Topoisomerase II and the microtubule dynamicity factor Stu2 / XMAP215 (see Figure 1). Failure to remove DNA intertwines causes anaphase chromatin bridges, and can lead to defects in chromosome segregation and thus, genomic instability and/or cell death.
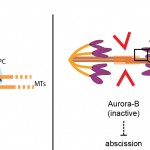
- What happens when chromatin bridges – such as those caused by Topoisomerase II defects – persist during cytokinesis? There are indications that a cytokinesis checkpoint, called NoCut, inhibits the last step of cell division (membrane abscission) when chromatin bridges are trapped in the cell cleavage site, therefore preventing DNA damage and ensuring genetic stability (see Figure 2). NoCut requires the kinase Aurora-B but its molecular mechanism remains elusive. We aim to understand how this new checkpoint works and what are the consequences of its inactivation in dividing cells. Some of the questions we need to answer are: What is the mechanism of abscission, and how is it regulated? Can premature abscission break lagging chromosomes? Anaphase chromatin bridges can have several distinct origins and properties – does the NoCut response depend on the type of chromatin bridge?